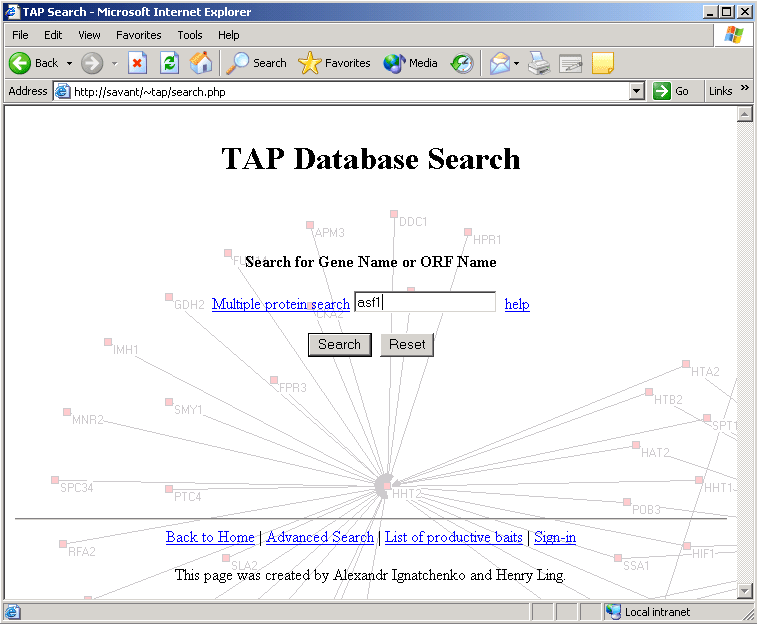
This page is divided into several sections:
As mentioned in the introduction page, present in this database are the results of the Tandem Affinity Purification (TAP) analysis of the S.Cerevisiae genome. At this site, the user can enter either the common name or the gene name of any gene in the S.Cerevisiae genome and all resulting interactors as determined through the TAP study will be listed. To begin with the most basic search, from the main search page the user can enter any single gene (in this example the query entered will be 'asf1').

This search will yield a page (called the interaction page) which will contain a list of proteins found to interact with the entered protein. This is a sample interaction page:
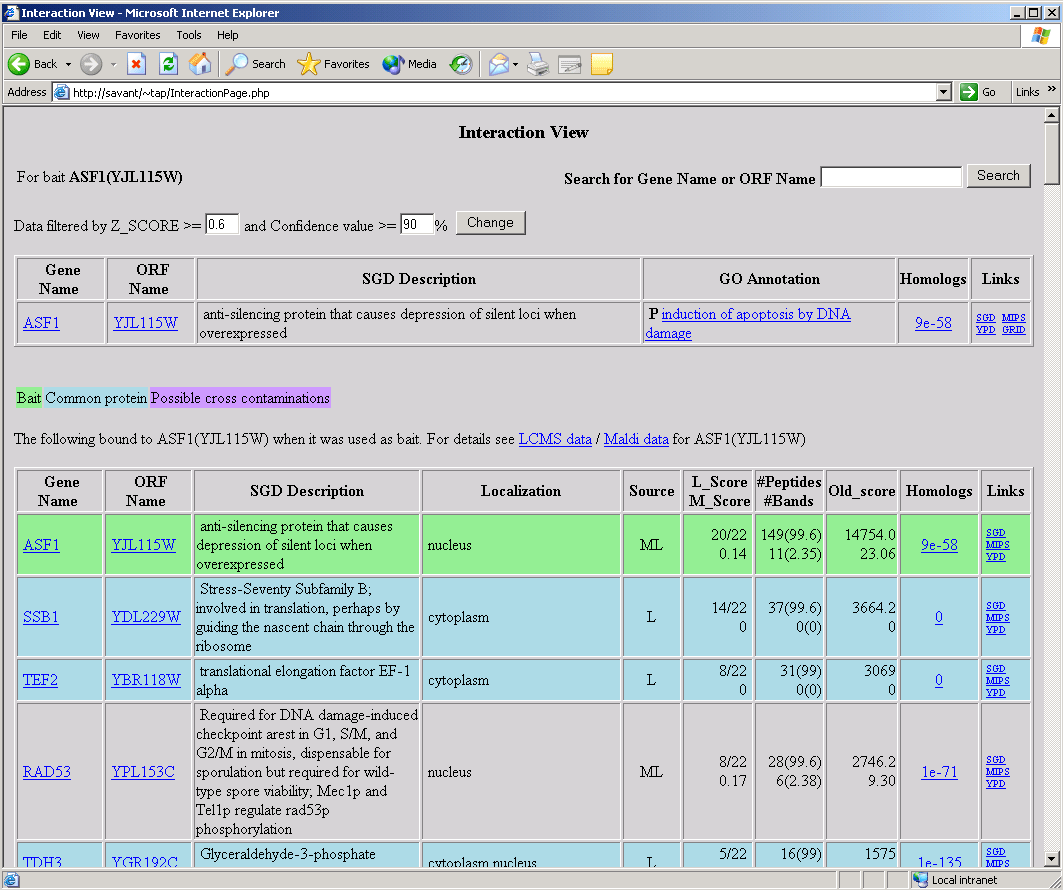
The bait name searched for will be indicated in the upper left corner of the screen. The default search parameters for Z-Score and confidence level can be altered in the boxes below the gene name and the search re-executed. The table just below this section gives such information as the gene name and description for the query gene/protein, and provides a link to the corresponding GO annotation.
The main feature of this page is a table containing all instances of interactions with the entered protein (or gene product). This table will appear twice on the interaction page, once for all instances where the queried protein bound to other proteins as 'bait' in TAP interactions, and again for the times when the queried protein acted as 'prey'.
The 'Source' column indicates whether the data was obtained by MALDI (M), LCMS (L) or both. Confidence scores for the respective testing method are given in the next few columns. The data obtained from either method can be obtained by clicking the corresponding hyperlink above the main table. Clicking the 'GFP' button in the 'Localization' column will generate a window containing a colored representation of the localization of the gene product (obtained from the UCSF database). Clicking the links in the 'Homologs' column will generate new browser windows containing lists of the homologs obtained by querying the SwissProt database. Clicking the hyperlinks in the 'Links' column will perform a search of the database selected using the gene featured in the corresponding row as a query. Clicking the links in the 'Gene Name' or 'ORF Name' columns will produce an interaction page generated with the selected item as the query. A new search can be initiated by entering the search item in the bar at the top right of the page, or by following the 'search' link at the bottom of the page.
Below the table are two buttons. The first button will submit the names of all of the genes in the table above it to Funspec. Funspec is a yeast cluster interpreter designed by this lab. It searches both public databases and published datasets and clusters inputted genes based on such criteria as localization, functional grouping and MIPS complex assignment. The second button will submit the same data to the GO term finder, which can retrieve information specifically on the function, cellular process of involvement, and cellular components of the inputted genes.